

International Journal of Agriculture, Environment and Biotechnology
Citation: IJAEB: 13(2): 145-148, June 2020
DOI: 10.30954/0974-1712.02.2020.4
BIOTECHNOLOGY
Molecular Characterization among Susceptible and Resistant Ridge Gourd Cultivars to Root-Knot Nematode, Meloidogyne incognita through ISSR Markers
ABSTRACT
The root-knot nematode, Meloidogyne incognita is the main nematode pest with host range more than 3000 plant species causes vast damage worldwide. The knowledge of genetic diversity in a crop species is essential for its improvement. Microsatellite sequences are especially suited to distinguish closely related genotypes in crop like ridge gourd. Several susceptible and resistant ridge gourd cultivars to root-knot nematode were identified on the basis of artificial screening and the present investigation was under taken for purity discrimination and characterize of these cultivars at molecular level taking ISSR markers. Standardized reported procedure was followed in order to conduct the experiment. The phylogenetic tree constructed by UPGMA method generated two main clusters i.e., cluster-I and cluster - II. The cluster I was further sub-divided into sub-cluster IA and IB. The cluster I consisted of three varieties (Aneeta, Aarti and BSS-1009), while cluster II consisted of two varieties (Harsha and Priya). Sub-cluster IA consisted of one variety (Aneeta), while Sub-cluster IB consisted of two varieties (Aarti and BSS-1009). Polymorphic per cent ranges from 13.16 per cent (between BSS-1009 and Aneeta ) to 43.18 per cent (between Aarti and Harsha). This study showed that ISSR markers produced specific DNA fragments for the identification of ridge gourd varieties. The knowledge acquired through this investigation may play a pivotal role in the application of molecular markers in ridge gourd improvement programmes.
Highlights
 Molecular characterization among susceptible and resistant ridge gourd cultivars to Meloidogyne incognita through ISSR Markers was studied in order to distinguish the cultivars with respect to banding patterns.
Molecular characterization among susceptible and resistant ridge gourd cultivars to Meloidogyne incognita through ISSR Markers was studied in order to distinguish the cultivars with respect to banding patterns.
Keywords: ISSR markers, Molecular characterization, Meloidogyne incognita, Ridge gourd cultivars
Ridge gourd (Luffa acutangula L.) is an important cucurbitaceae family vegetable crop with chromosome number 2n = 26. Ridge gourd is a tropical plant originated in India It is a annual, monoecious cross pollinated crop having green color fruit. The root-knot nematode, Meloidogyne incognita is the main nematode pest with host range more than 3000 plant species causes vast damage worldwide.
Molecular investigations are crucial for collection, conservation and its exploitation in future breeding programmes. The use of molecular marker for the evaluation of genetic diversity is receiving much attention than morphological markers. Microsatellite sequences are particularly suitable to differentiate closely related genotypes due to their high degree of variability therefore, favored in population studies (Smith and Devey 1994) and for classification of closely related cultivars (Vosman et al. 1992). The ISSR (Inter Simple Sequence Repeats) markers are DNA fragments of around 100 - 3000 bp located between adjacent, oppositely oriented microsatellite regions. The ISSR markers are amplified by PCR using microsatellite sequences as primers with a few selective nucleotides as anchors into the non-repeat adjacent regions (16 - 18 bp). Therefore, in the present experiment molecular characterization among ridge gourd genotypes were studied by using PCR based molecular markers for purity discrimination. Realizing the importance, present study on molecular characterization was done among susceptible and resistant ridge gourd cultivars to root-knot nematode using ISSR markers. The information acquired by this investigation might play an essential role in the application of molecular markers in ridge gourd improvement programmes.
MATERIALS AND METHODS
Several susceptible and resistant ridge gourd cultivars to root-knot nematode were identified on the basis of artificial screening and the present investigation was under taking five ridge gourd cultivars i.e., Priya and BSS -1009 (resistant), Aneeta (moderately resistant), Aarti and Harsha (highly susceptible) to root-knot nematode M. incognata. Total Genomic DNA extraction was done by following CTAB method developed by Rogers and Bendich (1994). Standardized reported RNAse procedure was followed in order to remove the RNA contaminants from DNA. The purity of DNA was further checked using NanoDrop ND-1000 spectrophometer. DNA samples with OD value 1.8 and 2.0 indicated the pure DNA free from proteins and carried forward for further proceeding. Four ISSR (Inter Simple Sequences Repeats) markers were used for the study. Sample ISSR PCR reaction mixture was prepared separately for each sample and primer combination by taking template DNA, Tag buffer, MgCl2, Taq DNA polymerase, dNTPs and primers. DNA amplification was done by PCR for 35 cycles, followed by Electrophoresis of ISSR PCR Products with 1.5 per cent agarose gel and 100 bp DNA ladder.
The DNA profile was visualized under UV (312 nm) trans-illuminator and documented using gel documentation system Gel DOCXRTM Imaging system UVP (USA). Clear and distinct bands
amplified by ISSR primers were scored for the presence (1) and absence (0) for the corresponding band among the cultivars. Data were analyzed and similarity matrix was constructed from binary data with Jaccard’s coefficients and dendrogram was constructed with un-weighted pair group method arithmatic average (UPGMA) algorithm, using Free Tree software (Hampl et al. 2001; Pavalice et al. 1999) and the dendrograms were visualized by Treeview 32 software (Page, 1996).
RESULTS AND DISCUSSION
The banding pattern of four ISSR markers in different susceptible and resistant ridge gourd cultivars to root-knot nematode are given in Table 1 and Fig. 1.
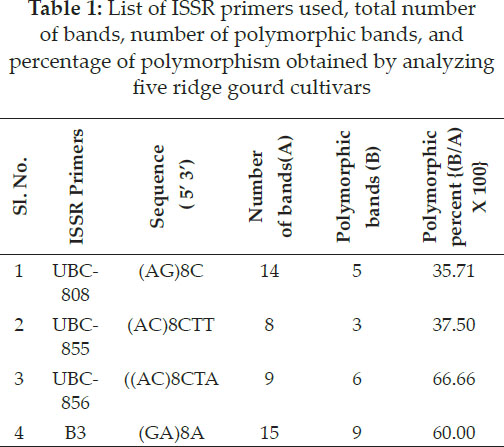
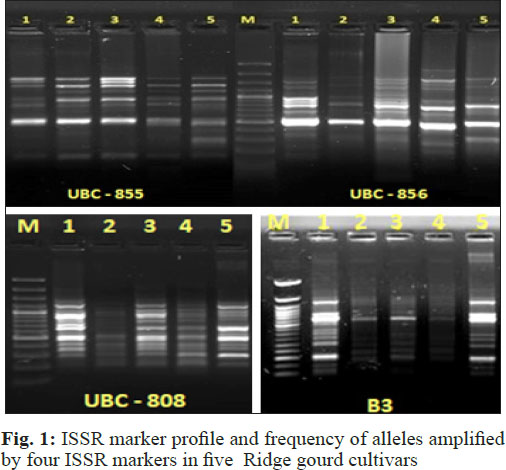
The pooled study of molecular marker through ISSR markers were used to confirm the similarity and differences between five ridge gourd varieties through Jaccard’s similarity coefficient and UPGMA method. Four ISSR primers generated total of 46 bands/alleles in which 23 bands were polymorphic. Primer B3 produced maximum 15 bands, while primer UBC-855 produced minimum 8 bands. The average bands per primer were 11.50. Maximum polymorphism observed for the primer UBC - 856 (66.66 per cent) and lowest for UBC - 808 (Table 1 and Fig. 1). Sikadar et al. (2010) observed that out of the ten (ISSR) primers, eight produced informative data for phylo-genetic analysis in Cucurbita species. These eight primers produced 139 ISSR fragments, an average of 17.37 bands per primer with amplified product sizes ranged from 599 to 2399 bp.
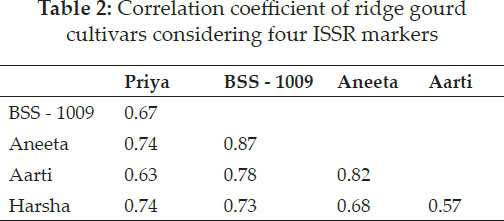
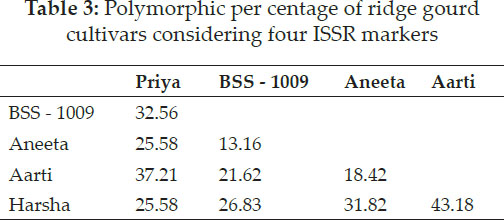
The Jaccard’s similarity coefficient and UPGMA method showed the highest (86.84 per cent) similarity between BSS - 1009 and Aneeta and the lowest (56.81 per cent) similarity between Aarti and Harsha (Table 2 and Table 3). Song et al. (2010) recorded that genetic similarity between wax gourd and chiehqua germplasm was in the range of 60 per cent to 99 per cent. Huang et al. (2010) observed that size of amplification ranged from 150 bp to 2700 bp with six ISSR primers in 38 diverse bitter gourd accessions. Resmi and Sreelathakumary (2011) estimated Jaccard’s similarity coefficients and constructed dendrogram by using UPGMA revealed the presence and extent of genetic similarities among the twenty-five landraces of ashgourd.
Pair-wise genetic similarities among the landraces determined using Jaccard’s coefficient ranged from 0.14 to 1.00.
Polymorphic per cent ranges from 13.16 per cent (between BSS - 1009 and Aneeta) to 43.18 per cent (between Aarti and Harsha). Latha (2012) studied RAPD analysis in cucumber and reported that OPA-19 primer recorded the highest polymorphism (86.6 per cent). Out of 130 bands, 115 bands were polymorphic for a specific primer and can be used as differential markers for varietal differentiation. Ravi et al. (2015) carried out for the comparison of RAPD and ISSR markers for polymorphism pattern and molecular diversity analysis among 17 ridge gourd (Luff a acutangula L.) and sponge gourd (Luff a cylindrica L.) genotypes using RAPD and ISSR markers. They reported out of 30 ISSR primers screened, twelve ISSR primers produced 79 bands of which 66 bands were polymorphic and 83.54 per cent polymorphism with an average of 7.16 bands per primer.
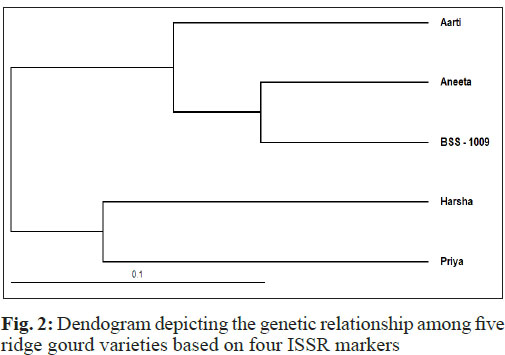
The ISSR data were used for UPGMA cluster analysis. The phylogenetic tree constructed by UPGMA method generated two main clusters i.e., cluster - I and cluster - II. The cluster I was further sub-divided into sub-cluster IA and IB (Fig. 2). The cluster I consisted of three varieties (Aneeta, Aarti and BSS - 1009), while cluster II consisted of two varieties (Harsha and Priya). Sub-cluster IA consisted of one variety (Aneeta), while Sub-cluster IB consisted of two cultivars (Aarti and BSS - 1009). Similar results have also been reported by Sikadar et al. (2010) and Huang et al. (2010). This study showed that ISSR markers produced specific DNA fragments for identification of ridge gourd varieties. De et al. (2015) studied phylo-genetic patterns andrelatedness among selected species of cucurbits using ISSR markers. They revealed a close genetic relationship between C. maderaspatanus and C. melo, while C. sativus, a member of the same genus, was placed as a distant relative from both species, thereby demonstrating remarkable diversification among members of the same genus. Chang et al. (2003) classified twenty pumpkin cultivars into three large categories and identified genetic distance of cluster ranging from 38 per cent and 100 per cent. Yildiz et al. (2014) characterized 24 accessions covering different groups of Cucumis melo L. by using 207 markers and found wide range of variations for investigated characteristics in Turkish gene pool that provides a good source of diversity to use in melon improvement program for better yield.
CONCLUSION
The phylogenetic tree constructed by UPGMA method generated two main clusters i.e., cluster - I and cluster - II. The cluster I was further subdivided into sub-cluster IA and IB. The cluster I consisted of three varieties (Aneeta, Aarti and BSS - 1009), while cluster II consisted of two varieties (Harsha and Priya). Sub-cluster IA consisted of one variety (Aneeta), while Sub-cluster IB consisted of two varieties (Aarti and BSS - 1009). Polymorphic per cent ranges from 13.16 per cent (between BSS - 1009 and Aneeta) to 43.18 per cent (between Aarti and Harsha). This study showed that ISSR markers produced specific DNA fragments for identification of ridge gourd varieties. The knowledge acquired through this investigation may play a pivotal role in the application of molecular markers in ridge gourd improvement programmes.
REFERENCES
Chang, H.B., Long, B., Park, Y., Jin Ma and Woojin, A. 2003. Assessment of genetic relationship among cucurbitaceae cultivars revealed by RAPD marker. Journal of Life Science, 13(5): 590-595.
De, P., Parab, M. and Singh, S. 2015. Article; agriculture and environmental biotechnology Intergenus variation analysis in few members of Cucurbitaceae based on ISSR markers. Biotechnology and Biotechnological Equipmen, 29(5): 882-886.
Hampl, V., Pavlicek A. and Flegr, J. 2001. Construction and bootstrap analysis of DNA fingerprinting based phylogenetic trees with the freeware program FreeTree: application to trichomonad parasites. International Journal of Systematic and Evolutionary Microbiology, 51: 731-735.
Huang, C.H., Wang, C.J. and Chyuan, J.H. 2010. Analysis of the genetic diversity and variety identifi cation of bitter gourd (Momordica charantia L.) by ISSR marker. Hualien District Agricultural Improvement Station, Bulletin, 28: 21-33.
Latha, K. 2012. Genetic diversity in 6 local cucumber varieties (Cucumis Satives) in karnataka market by RAPD-PCR technique. International Journal of Advanced Biological Research,2(1): 39-45.
Page, R.D. 1996.Tree View: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci., 12: 357-358.
Pavalice, A., Hrda, S. and Flegr, J. 1999. Free Tree-freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness, Application in the RAPD analysis of genus Frenkelia. Folia Biologica , 45: 97-99.
Ravi R. Rathod, Mehta, D.R., Gajera, H.P. and Delvadiya, N.A. 2015. Molecular characterization of ridge gourd (Luffa acutangula L.) and sponge gourd (Luffa cylindrica l.) genotypes through PCR based molecular markers. International Journal of Agriculture, Environment and Biotechnology, 8(3): 521-530.
Resmi, J. and Sreelathakumary, I. 2011. RAPD markers for genetic variability studies in ashgourd [Benincasa hispida (Thunb.) Cogn.]. Journal of Agricultural Technology, 7(4): 1097-110.
Rogers, S.O. and Bendich, A.J. 1994. Extraction of total cellular DNA from plants, algae and fungi; In: Gelvin, S.B. and Schilperoort, R.A. (eds.). Plant Molecular Biology manual, MA Kluwer Academic Publishers, Boston, pp. 1-8.
Sikadar, B., Bhaacharya, M., Mukherjee, A., Banerjee, A., Ghosh, E., Ghosh, B. and Roy, S.C. 2010. Genetic diversity in important members of Cucurabitaceae using isozyme, RAPD and ISSR markers. Biologia Plantarum, 54(1): 135-140.
Smith, N.D. and Devey, M.E. 1994. Occurrence and inheritance of microsatellites in Pinus radiata. Genomics, 37: 977-983.
Song, S.W., Li, Z., Liu, H.C., Sun, G.W. and Chen, R.Y. 2010. RAPD analysis of genetic diversity of wax gourd and chieh-qua germplasm. China Vegetables, 22: 47-53.
Vosman, M.I., Uzunova, M.I. and Ecke, W. 1992. Abundance, polymorphism and genetic mapping of microsatellites in Allium. Plant Breeding, 45: 156-167.
Yildiz, M., Akgul, N. and Sensoy, S. 2014. Morphological and molecular characterization of turkish landraces of Cucumis melo L. Notulae Botanicae Horti Agrobotanici, 42(1): 51-58.